Ongoing projects
My research explores the potential of combining my dual expertises in organic geochemistry and molecular biology.
1. Combined genomic and lipid analysis of 2-methylhopanes through Earth’s history
This project aims to constrain the utility of 2-methylhopanes as taxonomic biomarkers. 2-Methylhopanes were once hailed as promising cyanobacterial biomarkers and were extensively utilized to trace the evolution of oxygenic photosynthesis in the Proterozoic and even in the Archean. However, the subsequent discovery of 2-methylhopanoid production by non-cyanobacterial species called into question the diagnosticity of those compounds and further provoked debates on the general validity of biomarker studies in the Precambrian period.
I attempt to re-establish the diagnosticity of 2-methylhopanes by combining comprehensive genomic analyses of 2-methylhopanoid biosynthesis genes and non-contaminated up-to-date geochemical analyses of 2-methylhopanes in the geological record (see also the completed project 3 as described below).
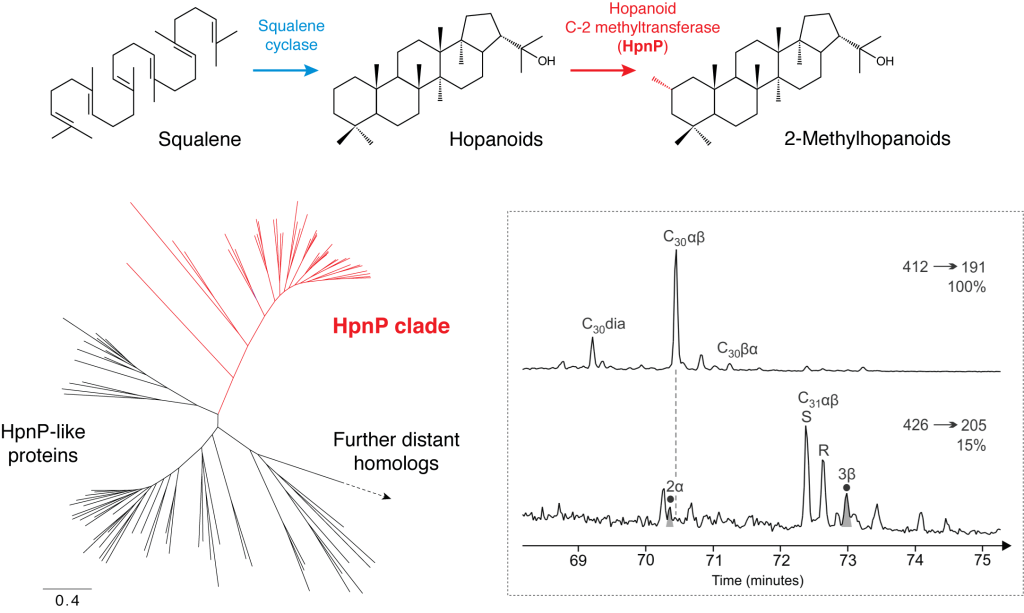
Fig. 2-Methylhopanoid biosynthesis by HpnP. In the geological record, 2-methylhopanoids are detected in the form of 2-methylhopanes (saturated forms of original compounds) as a result of diagenesis.
Collaborators
Jochen Brocks (Australian National University, Australia)
David Gold (University of California, Davis, USA)
References:
K. French, C. Hallmann, J. Hope, P. Schoon, J. Zumberge, Y. Hoshino, C. Peters, S. George, R. Buick, J. Brocks, R. Summons, 2015. Reappraisal of hydrocarbon biomarkers in Archean rocks, Proc. Natl. Acad. Sci. USA, 112, 5915-5920. (Geochemical Society, Best paper award, 2018) Link
P. Welander, M. Coleman, A. Sessions, R. Summons, & D. Newman (2010) Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes. Proc. Natl. Acad. Sci. USA 107(19):8537-8542. Link
2. Position-specific hydrogen isotope analysis for paleoclimatological reconstructions
The hydrogen isotopic ratio (D/H or δD) of plant- and algal-derived fossil lipids reflects the hydrogen isotopic composition of water used during photosynthesis and has been utilized as an essential paleohydrological proxy in paleoclimatology. However, our understanding of δD values in sedimentary lipid hydrocarbons is largely empirical and physiological and environmental variables are still poorly constrained.
This project applies position-specific isotope analysis (PSIA) to study D/H fractionation during lipid biosynthesis in order to quantify inherent biosynthetic fractionation effects from the environmental δD water signal and refine our understanding of the δD signal recorded in fossil lipid molecules. This will eventually allow for more precise and quantitative reconstructions of past hydrology, climate and metabolism (ecosystems).
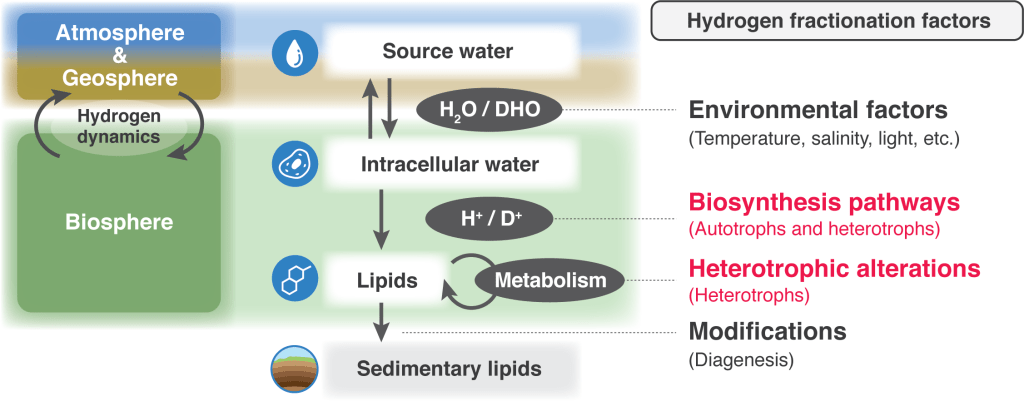
Fig. Interactions between the biosphere, geosphere and atmosphere. By better constraining isotope variation induced by lipid biosynthetic processes and addition of heterotrophic biomass (red color), we can refine the environmental information encoded in molecular D/H ratios.
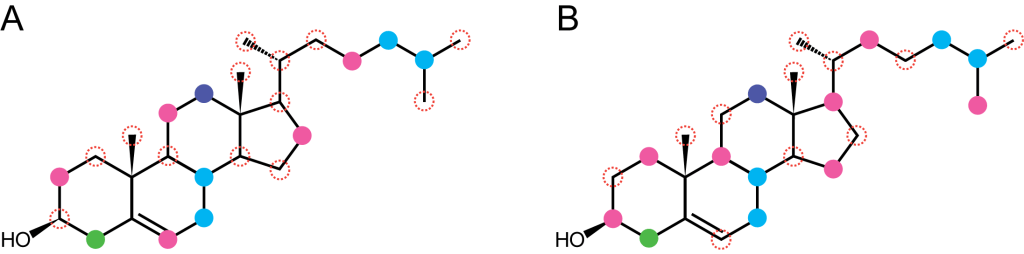
Fig. Isotopically-distinct cholesterol molecules that are produced by two different biosynthesis pathways. Colored dots indicate different biosynthetic sources. While bulk and compound-specific isotope analysis (CSIA) may not be able to distinguish the two versions of cholesterol, PSIA has a potential to discern them.
Reference
D. Sachse, I. Billault, G. Bowen, Y. Chikaraishi, T. Dawson, S. Feakins, K. Freeman, C. Magill, F. McInerney, M. van der Meer et al. (2012) Molecular Paleohydrology: Interpreting the Hydrogen-Isotopic Composition of Lipid Biomarkers from Photosynthesizing Organisms. Annual Review of Earth and Planetary Sciences, 40: 221-249. Link
3. Terpenoid biomarker analyses of land plant colonization (MAdLand project)
(DFG priority programme 2237)
Plant terrestrialization (water-to-land transition) occurred some 500 million years ago, but its detailed evolutionary trajectory remains enigmatic. This project aims to trace the early evolution of land plants in the geological record by the combination of comprehensive lipidomics analyses of early-diverging land plant species and lipid biomarker analyses in geological records from the Silurian and the Ordovician to the Cambrian.
This project is part of the bigger umbrella project MAdLand (Molecular Adaptation to Land: plant evolution to change).
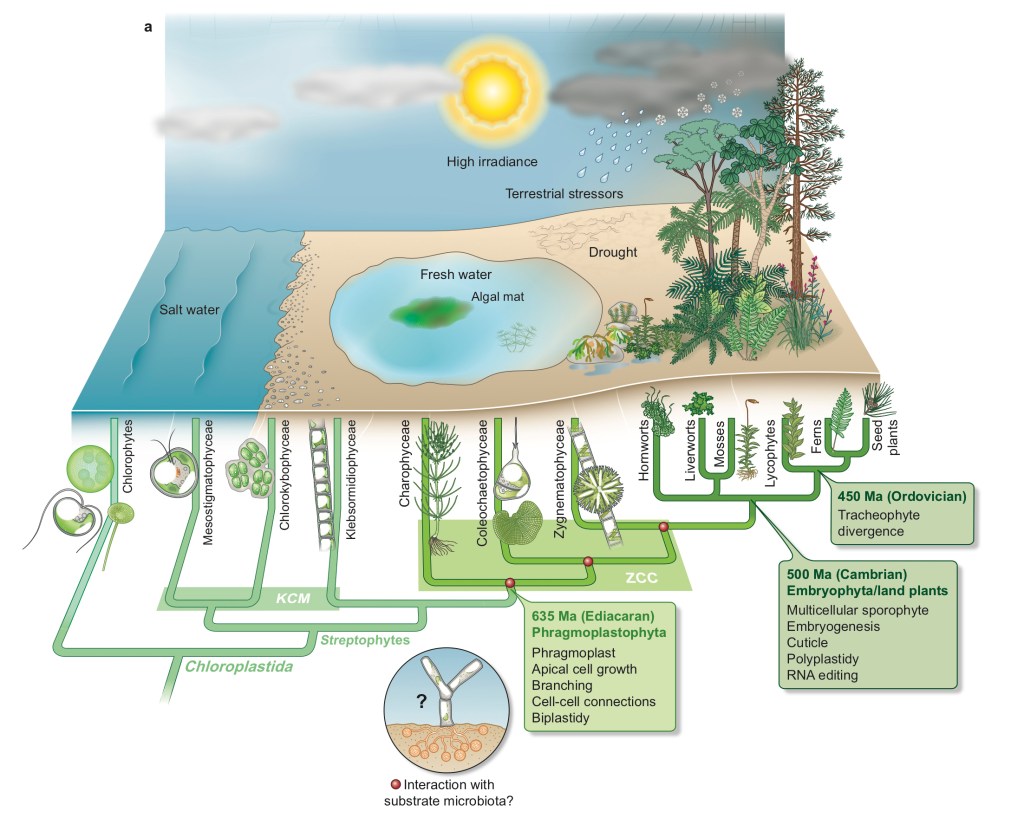
Fig. Proposed evolutionary history of Virdiplantae (Land plants & Chlorophytes; adapted from MAdLand website)
Collaborators
Sabine Zachgo (University of Osnabrueck, Germany)
Nicole Bale (NIOZ, Netherlands)
Reference:
T. Servais, B. Cascales-Miñana, C. Cleal, P. Gerrienne, D. Harper, M. Neumann (2019) Revisiting the Great Ordovician Diversification of land plants: Recent data and perspectives. Palaeogeography, Palaeoclimatology, Palaeoecology. 534:109280. Link
4. Evolution of eukaryotic cell wall biosynthesis
Eukaryotes produce a variety of distinct cell walls that enable host organisms to adapt to diverse environments. However, the evolutionary origin and the history of eukaryotic cell walls remain largely unsolved. In this project, comprehensive phylogenetic analyses are conducted for eukaryotic cell wall biosynthesis genes (in particular, glycosyltransferases for cellulose and chitin biosynthesis). In parallel, the taxonomical identity of cell wall remnants in the Precambrian rock record are analyzed through the comparison with modern eukaryotic cell walls.
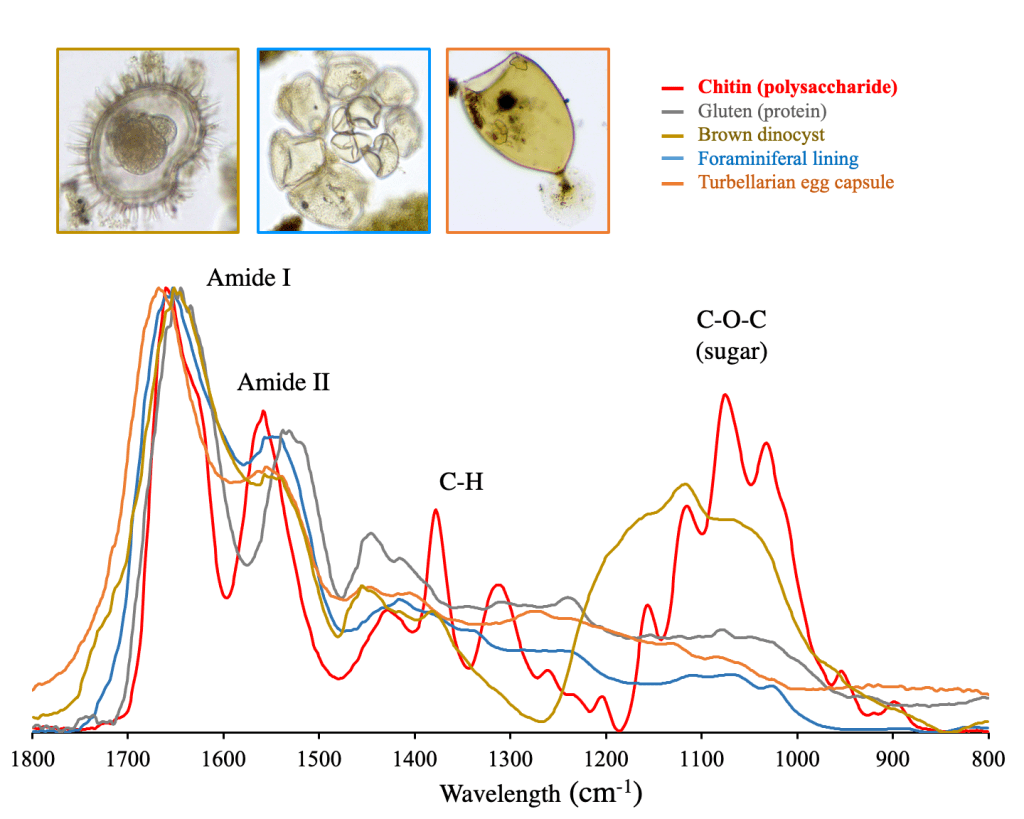
Fig. FT-IR measurements of modern eukaryotic cell walls.
Collaborator
Takuto Ando (Shimane University, Japan)
References
J. Fangel, P. Ulvskov, J. Knox, M. Mikkelsen, J. Harholt, Z. Popper, W. Willats (2012) Cell wall evolution and diversity. Frontiers in Plant Science, 3, 152. Link
Z. Popper, G. Michel, C. Hervé, D. Domozych, W. Willats, M. Tuohy, B. Kloareg, D. Stengel (2011) Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annual Review of Plant Biology, 62: 567-590. Link
Completed projects
1. Resurrection of ancestral steroid biosynthesis enzymes
(funded by Agouron Institute Geobiology Fellowship)
Eukaryogenesis (evolution of eukaryotes) is one of unsolved important questions in modern biology. Steroid biosynthesis is generally thought to be nearly exclusive to eukaryotes and thus steroids are extensively utilized as eukaryotic biomarkers in the geological record to constrain the evolutionary timing of eukaryogenesis. However, recent studies have increasingly suggested the presence of steroid biosynthesis in bacteria. Thus, addressing the evolutionary implication of bacterial steroid biosynthesis is of prime importance to correctly interpret the biological identity of steroids in the geological record and understand the evolutionary origin of eukaryotes.
In this project, comprehensive phylogenomic analyses of all steroid biosynthesis enzymes were conducted. Our results suggested the bacterial origin of steroid biosynthesis and the horizontal transfer of steroid biosynthesis genes from bacteria to eukaryotes, contrarily to the traditional understanding. Further, ancestral steroid-producing enzymes were experimentally resurrected, using the technique of Ancestral Sequence Reconstruction (ASR), and the catalytic products of resurrected enzymes were characterized to infer ancestral steroid compositions in early eukaryotes and bacteria. This project shed novel light on the process of eukaryogenesis through elucidating the evolutionary origin of steroid biosynthesis, which is thought to have been critical to form the eukaryotic-specific dynamic membrane system.
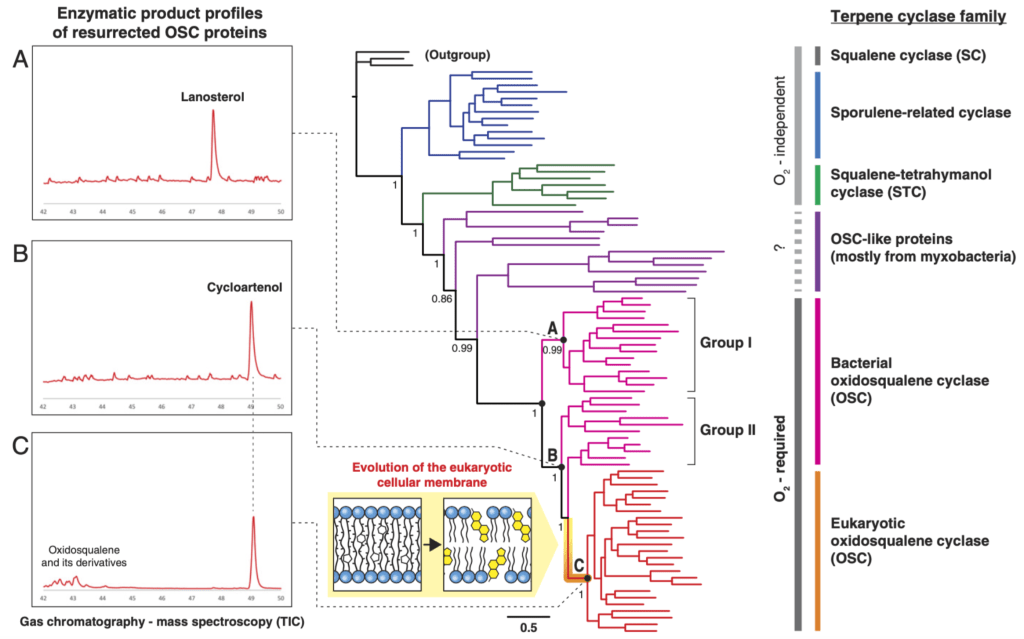
Fig. Phylogenetic tree of steroid-producing enzymes (oxidosqualene cyclase; OSC) and resurrection of ancestral OSC proteins (Nodes A, B & C).
References:
Y. Hoshino, E. Gaucher, 2021. Evolution of bacterial steroid biosynthesis and its impact on eukaryogenesis, Proc. Natl. Acad. Sci. USA, 118 (25) e2101276118. Link
Y. Hoshino, E. Gaucher, 2018. On the origin of isoprenoid biosynthesis, Molecular Biology and Evolution, 35, 2185-2197. Link
2. Neoproterozoic diversification of aerobic organisms
The evolution of complex multicellular organisms such as animals and plants is a hallmark of the aerobic evolution of life. However, the environmental setting that paved the way to this one of the most important evolutionary events on Earth remains enigmatic. This project examined the eukaryotic biomarker record (steranes) in the Cryogenian and the Ediacaran and discovered an abrupt and substantial shift of sterane composition that seems to reflect a drastic rearrangement of ecosystems at the terminal Proterozoic towards the onset of the Cambrian Explosion. Our results suggested that the primary production dominated by cyanobacteria for the most of the Proterozoic ended during the Cryogenian, followed by the rise of eukaryotic algae as the new major primary producers. The data particularly suggested the rise of green algae as the dominant producers. The project outcome substantially enhanced our understanding about the ecological evolution from ancestral cyanobacterial ecosystems towards modern algal-based ecosystems.
This project was only enabled by the establishment of non-contaminated sampling method for organic-lean Precambrian rocks as described in the completed project 3 below.
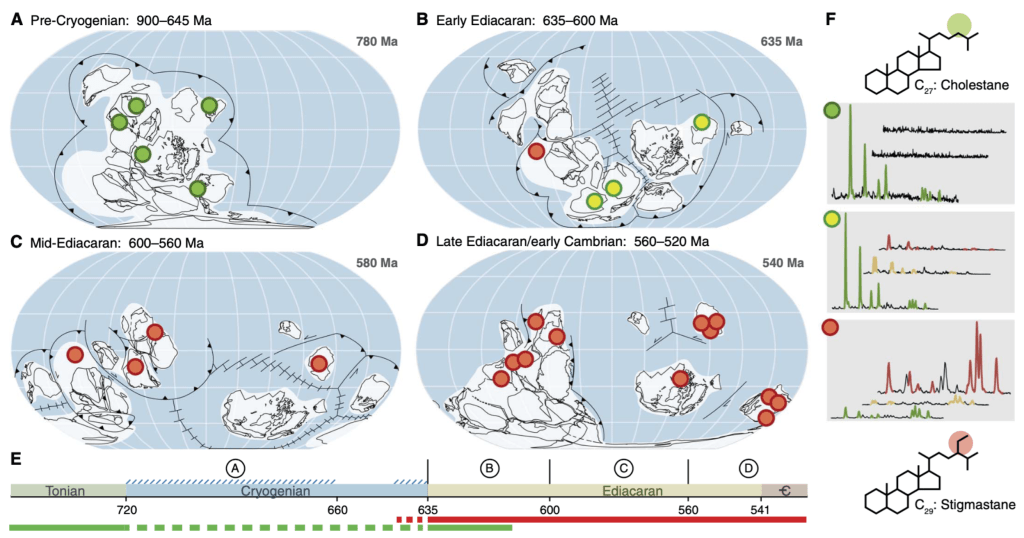
Fig. Sterane compositional shift from the Cryogenian to the Ediacaran (C27, C28 and C29 steranes).
Collaborators
Jochen Brocks (Australian National University, Australia)
David McKirdy (University of Adelaide, Australia)
Shuhai Xiao (Virginia Polytechnic Institute and State University, USA)
Supriyo Das (Presidency University, India)
Malgorzata Moczydlowska-Vidal (Uppsala University, Sweden)
References:
Y. Hoshino, A. Poshibaeva, W. Meredith, C. Snape, V. Poshibaev, G. et al. 2017. Cryogenian evolution of stigmasteroid biosynthesis, Science Advances, 3: e1700887. Link
J. Brocks, A. Jarrett, E. Sirantoine, C. Hallmann, Y. Hoshino, T. Liyanage, 2017. The rise of algae in Cryogenian oceans and the emergence of animals, Nature, 548, 578-581. Link
3. Re-assessment of Archean biomarker records
In this project, we developed an unprecedented hydrocarbon-clean protocol to collect and process Archean rock samples from the Pilbara region in Western Australia and demonstrated that once-considered authentic ‘Archean’ biomarkers are most likely contaminants that were introduced during sample handling. This project was performed by a consortium of five geochemistry labs in the world, including the Australian National University, Massachusetts Institute of Technology, University of California Riverside, Macquarie University, and the Max-Planck Institute for Biogeochemistry. As a positive corollary from the project outcome, the burden of proof for the detection of indigenous and syngenetic biomarkers throughout the Precambrian was significantly raised and led us to systematically (re-)analyze both previously studied and new Proterozoic sedimentary sequences with an unexpected outcome (see the ongoing project 1 and the completed project 2 described above).
In parallel to the drill core analyses, Archean outcrop samples were also collected across the Pilbara region and analyzed to evaluate the preservation potential of hydrocarbon biomarkers.
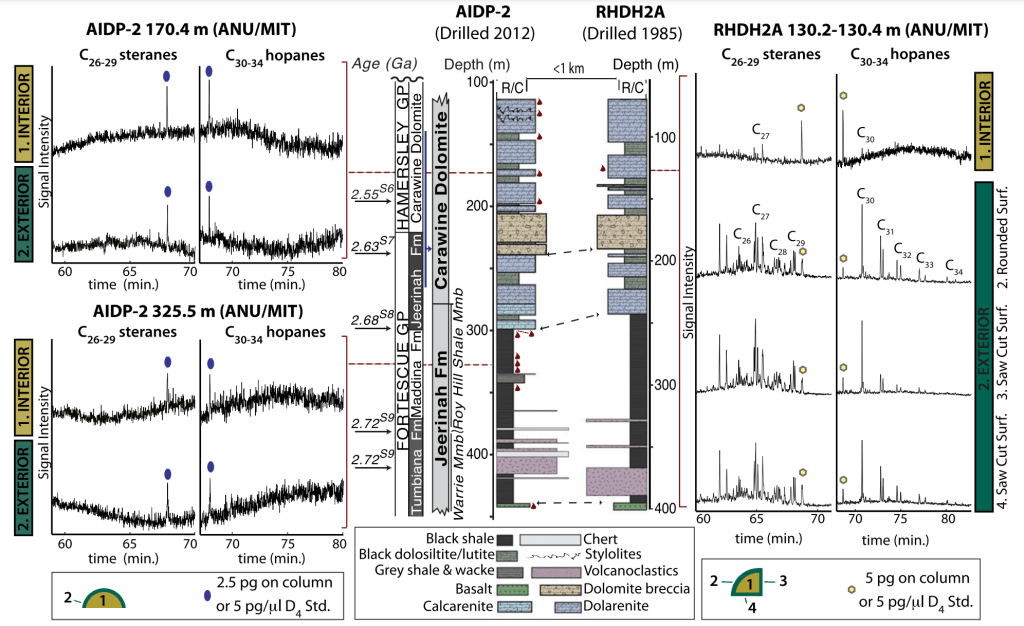
Fig. Non-contaminated Archean rock samples (left; AIDP-2) and previously-drilled contaminated Archean samples with substantial amounts of hopane and sterane biomarkers (right; RHDH2A).
Collaborators
Drill core analyses
Roger Summons (Massachusetts Institute of Technology, USA)
Christian Hallmann (Max-Planck Institute for Biogeochemistry, Germany)
Jochen Brocks (Australian National University, Australia)
Gordon Love (University of California Riverside, USA)
Outcrop analyses
David Flannery (Queensland University of Technology, Australia)
Malcolm Walter (University of New South Wales, Australia)
References
K. French, C. Hallmann, J. Hope, P. Schoon, J. Zumberge, Y. Hoshino, C. Peters, S. George, G. Love, R. Buick, J. Brocks, R. Summons, 2015. Reappraisal of hydrocarbon biomarkers in Archean rocks, Proc. Natl. Acad. Sci. USA, 112, 5915-5920. (Geochemical Society, Best paper award, 2018) Link
Y. Hoshino, S. George, 2015. Cyanobacterial inhabitation on the Archean rock surfaces in the Pilbara Craton, Western Australia, Astrobiology, 15, 559-574. Link
Y. Hoshino, D. Flannery, M. Walter, S. George, 2015. Investigation of hydrocarbon biomarkers preserved in an Archean rock at 2.7 Ga from the Fortescue Group in the Pilbara Craton in Western Australia, Geobiology, 13, 99-111. Link